
Andreas Martin
Molecular and Cell BiologyAndreas Martin is an Assistant Professor in the Department of Biochemistry, Biophysics and Structural Biology. His research goals are to decipher the fundamental principles that govern substrate engagement, deubiquitination, unfolding, and translocation by the eukaryotic 26S proteasome.
Spark Award Project
Jamming up Protein Turnover: Development of Specific Inhibitors for Deubiquitinating Enzymes
Reversible attachment of ubiquitin to proteins plays essential roles in protein trafficking, signaling, and degradation, and thus the control of many vital processes in eukaryotic cells. Important for this regulation is the rapid ubiquitin removal by deubiquitinating enzymes (DUBs), which therefore represent potent pharmacological targets for the treatment of cancers and neurodegenerative diseases. Protein degradation by the 26S proteasome critically depends on deubiquitination by the DUB Rpn11, a metalloprotease of the JAMM family. However, strategies for the selective inhibition of JAMM proteases are still missing. Our recent progress in recombinant expression, biochemical characterization, and structure-determination of Rpn11 offers unprecedented opportunities for the development of inhibitors for Rpn11 and related JAMM proteases by means of high-throughput screening. Specific activity assays will allow us to identify and validate new small-molecule antagonists, which are then tested for their effects on cancer-cell growth in established model systems. Identified inhibitors will be licensed to industry partners or used as the basis for starting a new company.
Andreas Martin’s Story
Too much of a good thing can be as bad as too little. It’s a lesson learned by many sunbathers, and it’s true for every cell in the body.

Proteins catalyze all metabolic reactions in cells. Others turn genes on and off, control the cell’s response to stress, or determine the pace of division. But each type of protein must do its job and then exit the scene, or risk killing the cell they serve.
In fact, cell survival depends as much on getting rid of proteins as producing them in the first place. Without an efficient disposal system, proteins clog the cell’s highly tuned machinery, throwing the cell into a death spiral.
The threat creates a novel strategy to treat cancer. Cancer cells produce an unusual amount of proteins at a rapid rate to support division, growth and invasion. As a result, they have a great need to rapidly degrade proteins, says Andreas Martin, assistant professor of biochemistry and biophysics.
Martin’s lab is launching a search for small-molecule drugs to block the ability of cancer cells to rid themselves of proteins. Doing so would overwhelm the cells with excess baggage and kill them. A successful drug would also expose cancer cells to tumor-suppressor proteins, which prevent rampant division.
And since cancer cells have a greater need than healthy cells to shed proteins, a drug that blocks the process would disproportionately threaten cancer cells, doing less harm to normal cells in the body.
Martin’s research focuses on crucial steps that all cells use to break down and recycle proteins. The natural process – only beginning to be well understood — relies on tens of thousands of barrel-shaped shredders known as proteasomes, which rove around each cell, ready to destroy any protein that has been earmarked for disposal.
The shredding process starts with a ubiquitous molecule called — reasonably — ubiquitin that tags proteins for destruction and presents them to the “input end” of the proteasome.
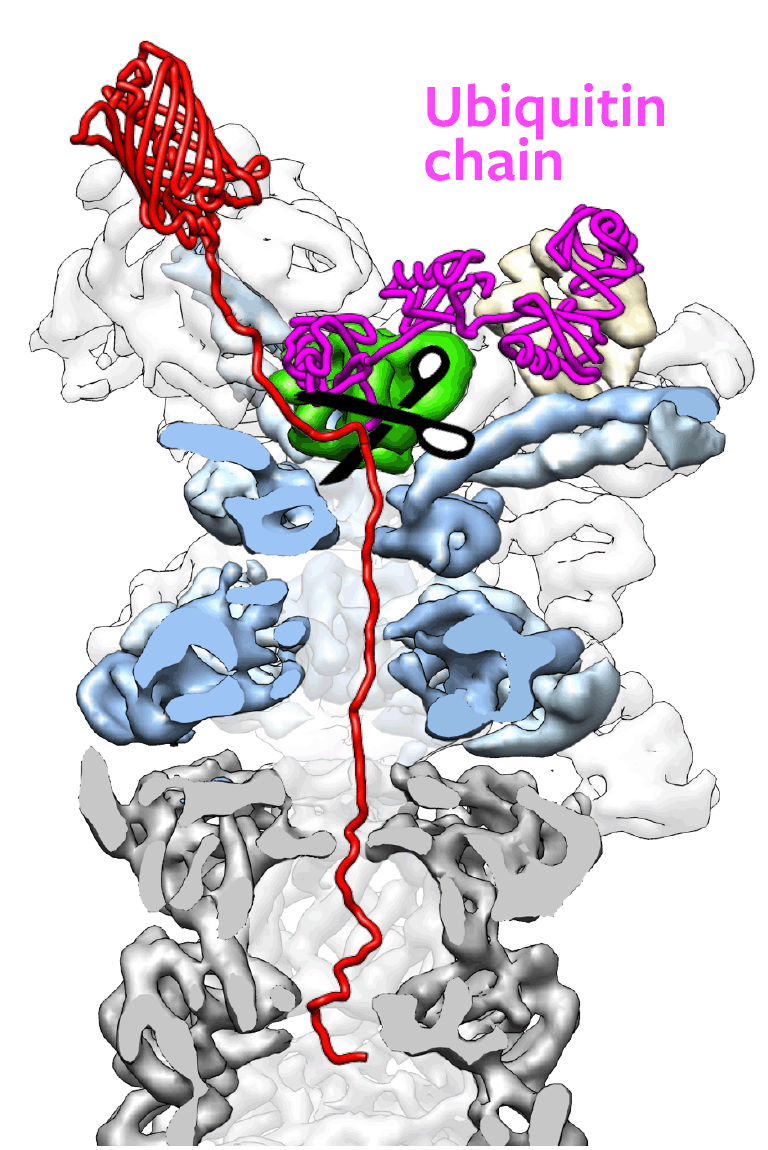
Ubiquitin must be snipped from a doomed protein before the proteasome machinery can pull that protein down into its maw. The snipping is done by a small part of the proteasome, called Rpn11 – the target that Martin seeks to attack. A drug that disables Rpn11 would leave ubiqutin and the protein locked together, shutting down the recycling process at its outset.
Martin plans to screen hundreds of thousands of compounds in search of an effective anti-proteasome drug. The research is supported by the Bakar Fellows Program, an initiative designed to accelerate advances from the lab to the marketplace. The funding will allow him to screen compounds at the UCSF-based Small Molecule Discovery Center, a kind of mini pharmaceutical research facility.
Several drugs that target and inhibit the proteasome are already in clinical use, but they affect processes farther down the protein-chewing pipeline. Martin’s focus on blocking one of the earliest steps should be more efficient and more selective, he says, causing fewer side effects in cancer treatment.
In 2012, Martin’s lab was able to determine the molecular architecture and precise location of all proteins that make up the bulky proteasome and play different roles during protein degradation.
The research relied in part on an imaging technique known as cryo-electron microscopy, which allows researchers to examine biological samples in their near-native physiological environment. This work, published in the journal Nature, led to determining Rpn11’s atomic-level structure and its mechanism of action — the key to finding a drug that might lock onto Rpn11 and disable it.
As part of the Bakar Fellows Program, Martin has ready access to entrepreneurs, patent lawyers, and investors who can provide advice and mentorship to help move his research toward commercialization.
“The Bakar support now enables us to translate our basic proteasome research into medical applications,” Martin says. “It’s the support we need to advance the research into developing novel anti-cancer drugs.”